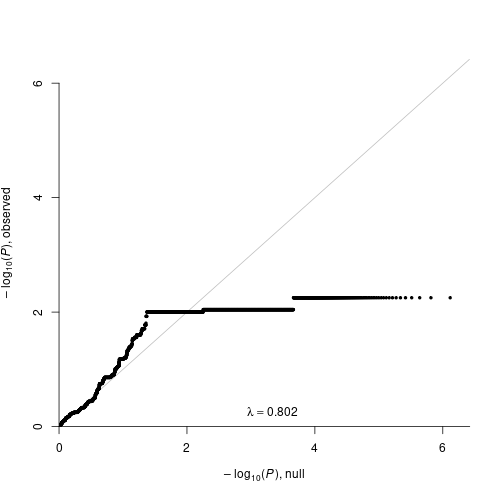
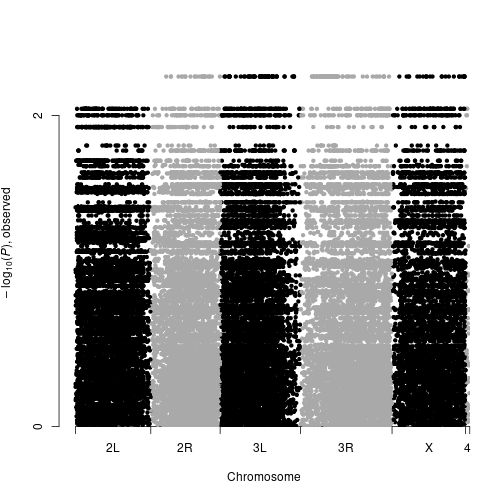
GWAS results were computed on dm3 genome.
If you want to make an independent analysis with DGRP2 or PLINK2, you can find here prepared input files for this phenotype:
DGRP2 results
PLINK2 results
Phenotype distribution and association to known covariates
Shapiro-Wilk test of normality (p-value=1.972e-2)
Null Hypothesis is rejected (p<=0.05).
Interpretation: NOT NORMAL distribution of this phenotype
Null Hypothesis is rejected (p<=0.05).
Interpretation: NOT NORMAL distribution of this phenotype
We tested for association of this phenotype with 6 known covariates Huang et al., 2014 using
both ANOVA and Kruskal-Wallis association tests.
ANOVA test
No significant result.
No significant result.
Kruskal-Wallis test
| Covariate | χ2 | p-value | Significance |
|---|---|---|---|
| wolba | 3.3409 | 0.0676 |